
| Source | Forward | Backward | Reversible |
|---|---|---|---|
| MetaCyc (?) Score of 1 represents the direction of the reaction as indicated in MetaCyc. | 0 | 0 | 1 |
| SEED (?) Score of 1 represents the direction of the reaction as indicated in the SEED database. | 0 | 0 | 1 |
| BiGG (?) Score represents 0.5 times the number of models with the indicated direction for this reaction in the BiGG database. | Not available | Not available | Not available |
| BRENDA (?) Score represents the proportion of reactions with the indicated direction reported in the BRENDA database for the enzymes associated with this reaction. Only reactions with a reported direction were used in the calculation. | 0.00 | 0.00 | 1.00 |
| Total (?) If no direction has >0.5 points, reaction is considered reversible. If the score of one direction with the highest score is more than 25% larger than the next highest score, the direction with the highest score is taken. Else, reaction is considered reversible. Exceptions (i.e., cases when these rules are overridden by other criteria) are idicated in general comments to the reaction. | 0.00 | 0.00 | 3.00 |
| Source | Forward | Backward | Reversible |
|---|---|---|---|
| MetaCyc (?) Score of 1 represents the direction of the reaction as indicated in MetaCyc. | 0 | 1 | 0 |
| SEED (?) Score of 1 represents the direction of the reaction as indicated in the SEED database. | 0 | 1 | 0 |
| BiGG (?) Score represents 0.5 times the number of models with the indicated direction for this reaction in the BiGG database. | Not available | Not available | Not available |
| BRENDA (?) Score represents the proportion of reactions with the indicated direction reported in the BRENDA database for the enzymes associated with this reaction. Only reactions with a reported direction were used in the calculation. | Not available | Not available | Not available |
| Total (?) If no direction has >0.5 points, reaction is considered reversible. If the score of one direction with the highest score is more than 25% larger than the next highest score, the direction with the highest score is taken. Else, reaction is considered reversible. Exceptions (i.e., cases when these rules are overridden by other criteria) are idicated in general comments to the reaction. | 0.00 | 2.00 | 0.00 |
| Source | Forward | Backward | Reversible |
|---|---|---|---|
| MetaCyc (?) Score of 1 represents the direction of the reaction as indicated in MetaCyc. | 1 | 0 | 0 |
| SEED (?) Score of 1 represents the direction of the reaction as indicated in the SEED database. | 1 | 0 | 0 |
| BiGG (?) Score represents 0.5 times the number of models with the indicated direction for this reaction in the BiGG database. | Not available | Not available | Not available |
| BRENDA (?) Score represents the proportion of reactions with the indicated direction reported in the BRENDA database for the enzymes associated with this reaction. Only reactions with a reported direction were used in the calculation. | 1.00 | 0.00 | 0.00 |
| Total (?) If no direction has >0.5 points, reaction is considered reversible. If the score of one direction with the highest score is more than 25% larger than the next highest score, the direction with the highest score is taken. Else, reaction is considered reversible. Exceptions (i.e., cases when these rules are overridden by other criteria) are idicated in general comments to the reaction. | 3.00 | 0.00 | 0.00 |
| Source | Forward | Backward | Reversible |
|---|---|---|---|
| MetaCyc (?) Score of 1 represents the direction of the reaction as indicated in MetaCyc. | Not available | Not available | Not available |
| SEED (?) Score of 1 represents the direction of the reaction as indicated in the SEED database. | Not available | Not available | Not available |
| BiGG (?) Score represents 0.5 times the number of models with the indicated direction for this reaction in the BiGG database. | Not available | Not available | Not available |
| BRENDA (?) Score represents the proportion of reactions with the indicated direction reported in the BRENDA database for the enzymes associated with this reaction. Only reactions with a reported direction were used in the calculation. | Not available | Not available | Not available |
| Total (?) If no direction has >0.5 points, reaction is considered reversible. If the score of one direction with the highest score is more than 25% larger than the next highest score, the direction with the highest score is taken. Else, reaction is considered reversible. Exceptions (i.e., cases when these rules are overridden by other criteria) are idicated in general comments to the reaction. | 0.00 | 0.00 | 0.00 |
| Source | Forward | Backward | Reversible |
|---|---|---|---|
| MetaCyc (?) Score of 1 represents the direction of the reaction as indicated in MetaCyc. | 1 | 0 | 0 |
| SEED (?) Score of 1 represents the direction of the reaction as indicated in the SEED database. | Not available | Not available | Not available |
| BiGG (?) Score represents 0.5 times the number of models with the indicated direction for this reaction in the BiGG database. | Not available | Not available | Not available |
| BRENDA (?) Score represents the proportion of reactions with the indicated direction reported in the BRENDA database for the enzymes associated with this reaction. Only reactions with a reported direction were used in the calculation. | Not available | Not available | Not available |
| Total (?) If no direction has >0.5 points, reaction is considered reversible. If the score of one direction with the highest score is more than 25% larger than the next highest score, the direction with the highest score is taken. Else, reaction is considered reversible. Exceptions (i.e., cases when these rules are overridden by other criteria) are idicated in general comments to the reaction. | 1.00 | 0.00 | 0.00 |
2.7.7.33Name: glucose-1-phosphate cytidylyltransferase Other names: CDP glucose pyrophosphorylase, cytidine diphosphoglucose pyrophosphorylase, cytidine diphosphate glucose pyrophosphorylase, cytidine diphosphate-D-glucose pyrophosphorylase, CTP:D-glucose-1-phosphate cytidylyltransferase click to see details on this item 4.2.1.45Name: CDP-glucose 4,6-dehydratase Other names: cytidine diphosphoglucose oxidoreductase, CDP-glucose 4,6-hydro-lyase click to see details on this item 1.17.1.1Name: CDP-4-dehydro-6-deoxyglucose reductase Other names: CDP-4-keto-6-deoxyglucose reductase, cytidine diphospho-4-keto-6-deoxy-D-glucose reductase, cytidine diphosphate 4-keto-6-deoxy-D-glucose-3-dehydrogenase, CDP-4-keto-deoxy-glucose reductase, CDP-4-keto-6-deoxy-D-glucose-3-dehydrogenase system, NAD(P)H:CDP-4-keto-6-deoxy-D-glucose oxidoreductase click to see details on this item 5.1.3.-Name: ascarylose synthesis click to see details on this item 1.1.1.-Name: ascarylose synthesis click to see details on this item |
[c] : g1pName: D-Glucose 1-phosphate Formula: C6H11O9P Charge: -2 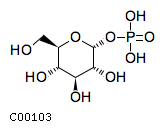 click to see details on this item + nadhName: Nicotinamide adenine dinucleotide - reduced Formula: C21H27N7O14P2 Charge: -2  click to see details on this item + nadphName: Nicotinamide adenine dinucleotide phosphate - reduced Formula: C21H26N7O17P3 Charge: -4  click to see details on this item + ctpName: CTP Formula: C9H12N3O14P3 Charge: -4  click to see details on this item + (2) hName: H+ Formula: H Charge: 1 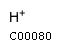 click to see details on this item |
 |
cdpasclName: CDP-ascarylose Formula: C15H23N3O14P2 Charge: -2  click to see details on this item + h2oName: H2O Formula: H2O Charge: 0  click to see details on this item + (2) piName: Phosphate Formula: HO4P Charge: -2 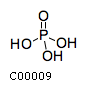 click to see details on this item + nadName: Nicotinamide adenine dinucleotide Formula: C21H26N7O14P2 Charge: -1  click to see details on this item + nadpName: Nicotinamide adenine dinucleotide phosphate Formula: C21H25N7O17P3 Charge: -3  click to see details on this item |
Unknown |